 |
D-Lib Magazine
November/December 2014
Volume 20, Number 11/12
Table of Contents
GROTOAP2 — The Methodology of Creating a Large Ground Truth Dataset of Scientific Articles
Dominika Tkaczyk, Pawel Szostek and Łukasz Bolikowski
Centre for Open Science, Interdisciplinary Centre for Mathematical and Computational Modelling, University of Warsaw, Poland
{d.tkaczyk, l.bolikowski}@icm.edu.pl; pawel.szostek@gmail.com
doi:10.1045/november14-tkaczyk
Printer-friendly Version
Abstract
Scientific literature analysis improves knowledge propagation and plays a key role in understanding and assessment of scholarly communication in scientific world. In recent years many tools and services for analysing the content of scientific articles have been developed. One of the most important tasks in this research area is understanding the roles of different parts of the document. It is impossible to build effective solutions for problems related to document fragments classification and evaluate their performance without a reliable test set, that contains both input documents and the expected results of classification. In this paper we present GROTOAP2 — a large dataset of ground truth files containing labelled fragments of scientific articles in PDF format, useful for training and evaluation of document content analysis-related solutions. GROTOAP2 was successfully used for training CERMINE — our system for extracting metadata and content from scientific articles. The dataset is based on articles from PubMed Central Open Access Subset. GROTOAP2 is published under Open Access license. The semi-automatic method used to construct GROTOAP2 is scalable and can be adjusted for building large datasets from other data sources. The article presents the content of GROTOAP2, describes the entire creation process and reports the evaluation methodology and results.
Keywords: Document Content Analysis, Zone Classification, System Evaluation, Ground Truth Dataset
1. Introduction
The analysis of scientific literature accelerates spreading ideas and knowledge and plays a key role in many areas of research, such as understanding scholarly communication, the assessment of scientific results, identifying important research centers or finding interesting unexplored research possibilities. Scientific literature analysis supports a number of tasks related to metadata and information extraction, scientific data organizing, providing intelligent search tools, finding similar and related documents, building citation networks, and many more. One of the most important tasks in this research area is understanding the roles of different fragments of documents, which is often referred to as document zone or block classification. An efficient zone classification solution needs to be carefully evaluated, which requires a reliable test set containing multiple examples of input documents and the expected results of classification.
Even narrowed to analysing scientific publications only, document zone classification is still a challenging problem, mainly due to the vast diversity of document layouts and styles. For example, a random subset of 125,000 documents from PubMed Central contains scientific publications from nearly 500 different publishers, many of which use original layouts and styles in their articles. Therefore it is nearly impossible to develop a high quality, generic zone classification solution without a large volume of ground truth data based on a diverse document set.
The role played by a given document fragment can be deduced not only from its text content, but also from the way the text is displayed for the readers. As a result, document zone classifier can benefit a lot from using not only the text content of objects, but also geometric features, such as object dimensions, positions on the page and in the document, formatting, object neighbourhood, distance between objects, etc. A dataset useful for training such a classifier must therefore preserve the information related to object size, position and distance.
In this paper we present GROTOAP2 (GROund Truth for Open Access Publications) — a large and diverse test set built upon a group of scientific articles from PubMed Central Open Access Subset. GROTOAP2 contains 13,210 ground truth files representing the content of publications in the form of a hierarchical structure. The structure stores the full text of a publication's source PDF file, preserves the geometric features of the text, and also contains zone labels. The corresponding PDF files are not directly included in the dataset, but can be easily obtained online using provided scripts. GROTOAP2 is distributed under the CC-BY license in the Open Access model and can be downloaded here.
The method used to create GROTOAP2 is semi-automatic and requires a short manual phase, but does not include manual correction of every document and therefore can scale easily to produce larger sets. The method can also be adapted to produce similar datasets from other data sources.
GROTOAP2 test set is very useful for adapting, training and performance evaluation of document analysis-related solutions, such as zone classification. The test set was built as a part of the implementation of CERMINE [8] — a comprehensive open source system for extracting metadata and parsed bibliographic references from scientific articles in born-digital form. GROTOAP2 has been successfully used for training and performance evaluation of CERMINE's extraction process and its two zone classifiers. The whole system is available here.
In the rest of the paper we present the content of GROTOAP2, compare our solution to existing ones and discuss its advantages and drawbacks. We also describe in details the semi-automatic process of creating the dataset, report the evaluation methodology and results.
2. Previous Work
Existing test sets containing ground truth data useful for zone classification are usually based on scanned document images instead of born-digital documents. For example UW-III contains various document images along with structure-related ground truth information. Unfortunately UW-III is not free and difficult to purchase. MARG is a dataset containing scanned pages from biomedical journals. The main problem is that it contains only the first pages of publications and only a small subset of zones is included, and as a result its usability for performance evaluation of page segmentation and zone classification is very limited. PRImA [1] dataset is also based on document images of various types and layouts, not only scientific papers. Other data sets built upon scanned document images of various layouts are: MediaTeam Oulu Document Database [5], UvA dataset and Tobacco800 [3].
Open Access Subset of PubMed Central contains around 500,000 life sciences publications in PDF format, as well as their metadata in associated NLM files. NLM files contain a rich set of metadata of the document (title, authors, affiliations, abstract, journal name, etc.), full text (sections, headers and paragraphs, tables and equations, etc.), and also document's references with their metadata. This makes PMC a valuable set for evaluating document analysis-related algorithms. Unfortunately, provided metadata contains only labelled text content and lacks the information related to the way the text is displayed in the PDF files.
GROTOAP2 is a successor of GROTOAP [7], a very similar, but much smaller, semi-automatically created test set. GROTOAP was created with the use of zone classifiers provided by CERMINE. First, a small set of documents was labelled manually and used to train the classifiers. Then, a larger set was labelled automatically by retrained classifiers and finally the results were corrected by human experts. Since GROTOAP's creation process required a manual correction of every document by an expert, the resulting test set is relatively small and every attempt to expand it is time-consuming and expensive. Due to the small size and lack of diversity, algorithms trained on GROTOAP did not generalize well enough and performed worse on diverse sets.
In GROTOAP2 all these drawbacks were removed. The dataset is based on born-digital PDF documents from PMC and preserves all the geometric features of objects. In contrast to GROTOAP, the method used to construct GROTOAP2 is scalable and efficient, which allowed for constructing much larger and diverse dataset. Table 1 compares the basic parameters of GROTOAP and GROTOAP2 and shows the difference in size and diversity.
| Number of: |
GROTOAP |
GROTOAP2 |
| publishers |
12 |
208 |
| documents |
113 |
13,210 |
| pages |
1,031 |
119,334 |
| zones |
20,121 |
1,640,973 |
| zone labels |
20 |
22 |
Table 1: The comparison of the parameters of GROTOAP and GROTOAP2 datasets. The table shows the numbers of different publishers included in both datasets, as well as the numbers of documents, pages, zones and zone labels.
3. GROTOAP2 Dataset
GROTOAP2 is based on documents from PubMed Central Open Access Subset. The dataset contains:
- 13,210 ground-truth files in XML format holding the content of scholarly publications in hierarchical structured form, which preserves the entire text content of the article, the geometric features related to how the text is displayed in the corresponding PDF file, and also the labels for text zones.
- A list of URLs of corresponding scientific articles in PDF format.
- A bash script, that can be used to download PDF files from PMC repository.
The dataset contains 13,210 documents with 119,334 pages and 1,640,973 zones in total, which gives the average of 9.03 pages per document and 13.75 zones per page. GROTOAP2 is a diverse dataset and contains documents from 208 different publishers and 1,170 different journals. Table 2 and Figure 1 show the most popular journals and the most popular publishers included in the dataset.
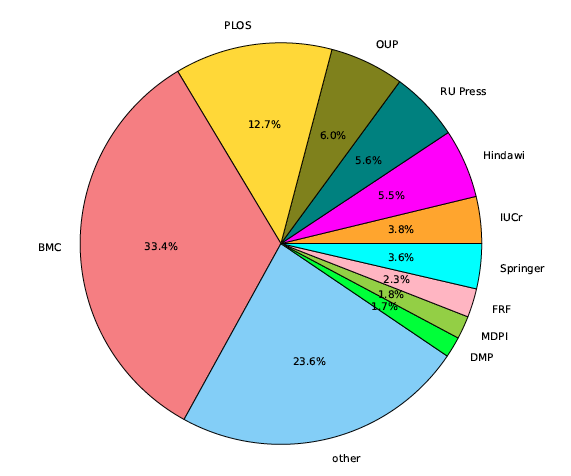
Figure 1: The most popular publishers included in GROTOAP2 test set. The abbreviations stand for: BMC — BioMed Central, PLOS — Public Library Of Science, OUP — Oxford University Press, RU Press — The Rockefeller University Press, Hindawi — Hindawi Publishing Corporation, IUCr — International Union of Crystallography, Springer — Springer Verlag, FRF — Frontiers Research Foundation, MDPI — Molecular Diversity Preservation International, DMP — Dove Medical Press
| Journal |
Number of articles |
Fraction of the dataset |
| PLOS ONE |
1212 |
9.17% |
| Nucleic Acids Research |
575 |
4.35% |
| Acta Crystallographica |
440 |
3.33% |
| The Journal of Cell Biology |
386 |
2.92% |
| The Journal of Experimental Medicine |
234 |
1.77% |
| BMC Public Health |
201 |
1.52% |
| Journal of Medical Case Reports |
169 |
1.28% |
| BMC Cancer |
152 |
1.15% |
| BMC Genomics |
147 |
1.11% |
| PLOS Genetics |
114 |
1.09% |
Table 2: The most popular journals in GROTOAP2 test set. The table shows how many documents from each journal are included in the dataset as both the exact number and the fraction of the dataset.
3.1 Ground truth structure
The main part of GROTOAP2 are ground truth files built from scholarly articles in PDF format. A ground truth file contains a hierarchical structure that holds the content of an article preserving the information related to the way elements are displayed in the corresponding PDF file.
The hierarchical structure represents an article as a list of pages, each page contains a list of zones, each zone contains a list of lines, each line contains a list of words, and finally each word contains a list of characters. Each structure element can be described by its text content, position on the page and dimensions. The structure stored in a ground truth file contains also the natural reading order for all structure elements. Additionally, labels describing the role in the document are assigned to zones.
The smallest elements in the structure are individual characters. A word is a continuous sequence of characters placed in one line with no spaces between them. Punctuation marks and typographical symbols can be separate words or parts of adjacent words, depending on the presence of spaces. Hyphenated words that are divided into two lines appear in the structure as two separate words that belong to different lines. A line is a sequence of words that forms a consistent fragment of the document's text. Words placed geometrically in the same line of the page, that are parts of neighbouring columns, do not belong to the same line. A zone is a consistent fragment of the document's text, geometrically separated from surrounding fragments and not divided into paragraphs or columns.
All bounding boxes are rectangles with edges parallel to the page's edges. A bounding box is defined by two points: left upper corner and right lower corner of the rectangle. The coordinates are given in typographic points (1 typographic point equals to 1/72 of an inch). The origin of the coordinate system is the left upper corner of the page.
Every zone is labelled with one of 22 labels:
- abstract — document's abstract, usually one or more zones placed in the first page of the document;
- acknowledgments — sections containing information about document's acknowledgments or funding;
- affiliation — authors' affiliations sections, which sometimes contain contact information (emails, addresses) as well;
- author — a list of document's authors,
- bib_info — zones containing all kinds of bibliographic information, such as journal/publisher name, volume, issue, DOI, etc.; often includes pages' headers or footers; headers and footers containing document's title or author names are also labelled as bib_info;
- body_content — the text content of the document, contains paragraphs and section titles;
- conflict_statement — conflict statement declarations;
- copyright — copyright or license-related sections;
- correspondence — the authors' contact information, such as emails or addresses;
- dates —important dates related to the document, such as received, revised, accepted or published dates;
- editor — the names of the document's editors;
- equation — equations placed in the document;
- figure — zones containing figures' captions and other text fragments belonging to figures;
- glossary — important terms and abbreviations used in the document;
- keywords — keywords listed in the document;
- page number —zones containing the numbers of pages;
- references —all fragments containing bibliographic references listed in the document;
- table — the text content of tables and table captions;
- title — the document's title;
- title_author — zones containing both document's title and the list of authors;
- type —the type of the document, usually mentioned on the first page near the title, such as "research paper", "case study" or "editorial";
- unknown — used for all the zones that do not fall in any of the above categories.
A zone that contains only a title of a section, such as "Abstract", "Acknowledgments" or "References" is labelled with the same label as the section itself. Figure 2 shows fractions of the documents from the dataset that contain a given label.
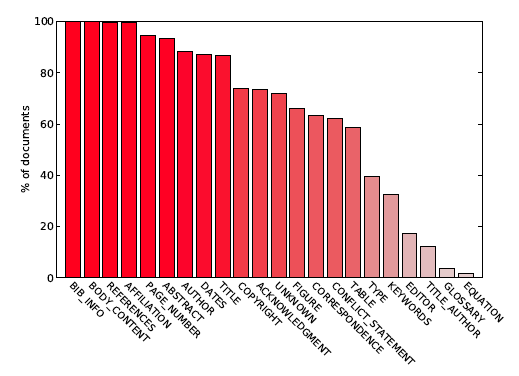
Figure 2: The diagram lists the labels in GROTOAP2 dataset, and for each label shows the fraction of documents in the dataset that contain zones with a given label.
3.2 Ground truth file format
We use TrueViz format [2] for storing ground truth files. TrueViz is an XML-based format that allows to store the geometrical and logical structure of the document, including pages, zones, lines, words, characters, their content and bounding boxes, and also zone labels and the order of the elements. The listing below shows a fragment of an example ground truth file from GROTOAP2. Repeated fragments or fragments that are not filled have been omitted.
XML-based format makes ground truth files easily readable by machines, but also makes them grow to enormous size, much greater than related PDFs. To limit the size of the dataset, only ground truth files were included. The corresponding PDF files can be easily downloaded using provided URL list or directly using a bash script.
4. The Method of Building GROTOAP2 Dataset
GROTOAP2 dataset was created semi-automatically from PubMed Central resources (Figure 3):
- First, a large set of files was downloaded from Open Access Subset of PubMed Central. We obtained both source articles in PDF and corresponding NLM files containing metadata, full text and references.
- PDF files were automatically processed by tools provided by CERMINE and their hierarchical geometric structure along with the natural reading order was constructed.
- The text content of every extracted zone was compared to labelled data from corresponding NLM files, which resulted in attaching the most probable labels to zones.
- Files containing a lot of zones with unknown labels, that is zones for which the automatic labelling process was unable to determine the label, were filtered out.
- A small random sample of the remaining documents was inspected by a human expert. We were able to identify a number of repeated problems and errors and develop heuristic-based rules to correct them.
- From the corrected files the final dataset was chosen randomly.
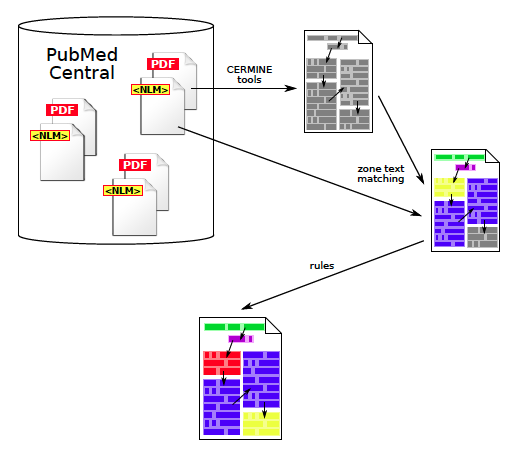
Figure 3: The process of creating GROTOAP2 dataset. First, PDF files from PMC were processed in order to extract characters, words, lines and zones. Then, the text content was matched against annotated NLM files from PMC, which resulted in zone labelling. Finally, some of the most often repeated errors were removed by simple rules.
4.1 Ground truth generation
PDF and NLM files downloaded from PubMed Central Open Access Subset were used to generate the geometric hierarchical structures of the publications' content, which were stored using TrueViz format [2] in ground truth files.
In the first phase of ground truth generation process we used automatic tools provided by CERMINE [8]. First, the characters were extracted from PDF files. Then, the characters were grouped into words, words into lines and finally lines into zones. After that reading order analysis was performed resulting in elements at each hierarchy level being stored in the order reflecting how people read manuscripts.
In the second phase the text content of each zone extracted previously was matched against labelled fragments extracted from corresponding NLM files. We used Smith-Watermann sequence alignment algorithm [6] to measure the similarity of two text strings. For every zone, a string with the highest similarity score above a certain threshold was chosen from all strings extracted from NLM. The label of the chosen string was then used to assign a functional label to the zone. If this approach failed, the process tried to use "accumulated" distance, which makes it possible to assign a label to these zones that form together one NLM entry, but were segmented into several parts. If none of the similarity scores exceeded the threshold value, the zone was labelled as unknown. After processing the entire page, an additional attempt to assign a label to every unknown zone based on the labels of the neighbouring zones was made.
4.2 Filtering files
Data in NLM files vary greatly in quality from perfectly labelled down to containing no valuable information. Poor quality NLMs result in sparsely labelled zones in generated TrueViz files, as the labelling process has no data to compare the zone text content to. Hence, it was necessary to filter documents whose zones are classified in satisfying measure. Figure 4 shows a histogram of documents with specified percentage of zones labeled with concrete classes. There are many documents (43%) having more than 90% of zones labelled, and only those documents were selected for further processing steps.
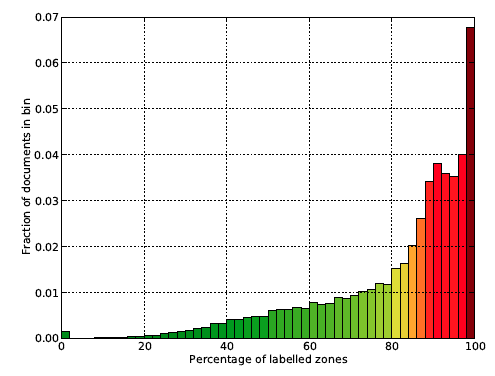
Figure 4: Histogram of documents in the dataset having given percentage of zones with an assigned class value.
We also wanted to be sure that the layout distributions in the entire processed set and the selected subset are similar. The layout distribution in a certain document set can be approximated by publisher or journal distribution. If poor quality metadata was associated with particular publishers or journals, choosing only highly covered documents could result in eliminating particular layouts, which was to be avoided. We calculated the similarity of publisher distributions of two sets using the following formula:
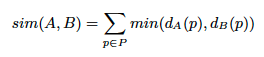
where P is the set of all publishers in the dataset and dA(p) and dB(p) are the percentage share of a given publisher in sets A and B, respectively. The formula yields 1.0 for identical distributions, and 0.0 in the case of two sets, which do not share any publishers. The same formula can be used to calculate the similarity with respect to journal distribution. In our case the similarity of publisher distributions of the entire processed set and a subset of documents with at least 90% labelled zones is 0.78, and the similarity of journal distributions 0.70, thus the distributions are indeed similar.
4.3 Ground truth improvement
After filtering files we randomly chose a sample of 50 documents, which were subjected to a manual inspection done by a human expert. The sample was big enough to show common problems occurring in the dataset, and small enough to be manually analyzed within a reasonable time. The inspection revealed a number of repeated errors made by the automatic annotation process. At this point we decided to develop a few simple heuristic-based rules, which would significantly reduce the error rate in the final dataset. Some examples of the rules are:
- In some cases the zone labelled as title contained not only the title as such, but also a list of document's authors. We decided to introduce a new label title_author for such cases. We detect those zones based on the similarity measures and assign the new label to them.
- Pages numbers included in NLM files are usually page ranges from the entire journal volume or conference proceedings book. In PDF file, on the other hand, pages are sometimes simply numbered starting from 1. In such cases the zones containing page numbers could not be correctly labelled using text content matching. We detect those zones based on their text content and the distance to the top or bottom on the page and label them as page_number.
- Figures' captions were often mislabelled as body_content, especially when the caption contains very similar text to one of the paragraphs. We used a regexp to detect such zones and label them as figure.
- Similarly, tables' captions mislabelled as body_content are recognized by a regexp and labelled as table.
- A table placed in the document often contains many small zones, especially if every cell forms a separate zone. The text content of such a small zone can often be found in document's paragraphs as well, therefore those zones are sometimes mislabelled as body_content. To correct this, we detect small zones lying in the close neighbourhood of table zones and label them as table.
- Due to the lack of data in NLM files, zones containing information related to document's editors, copyright, acknowledgments or conflict statement are sometimes not labelled or mislabelled. Since those zones are relatively easy to detect based on section titles and characteristic terms, we use regexps to find them and label them correctly.
- Zones that occur on every page or every odd/even page and are placed close to the top or bottom of the page were often not labelled, in such cases we assigned bib_info label to them.
The inspection also revealed segmentation problems in a small fraction of pages. The most common issue were incorrectly constructed zones, for example when the segmentation process put every line in a separate zone. Those errors were also corrected automatically by joining the zones that are close vertically and have the same label.
From the dataset improved by the heuristic-based rules we randomly chose 13,210 documents for the final set.
4.4 Application to other datasets
The process can be reused to construct a dataset containing labelled zones from a different set of documents. It will need source document files in PDF format and any form of annotated textual data that can be matched against the sources' content.
In the reused ground truth generation process the automatic matching step can be used out of the box, while the manual inspection step has to be repeated and possibly a different set of rules have to be developed and applied. Fortunately, the process does not require manual correction of every document, and scales well even to very large collections of data.
Proposed method cannot be applied if there is no additional annotated data available. In such cases the dataset can be built in a non-scalable way using a machine learning-based classifier, similarly to the way the first version of GROTOAP was created.
5. Evaluation
The process of creating GROTOAP2 did not include manual inspection of every document by a human expert, which allowed us to create a large dataset, but also caused the following problems:
- segmentation errors resulting in incorrectly recognized zones and lines and their bounding boxes,
- labeling errors resulting in incorrect zone labels.
GROTOAP2 dataset was evaluated in order to estimate how accurate the labelling in the ground truth files is. Two kinds of evaluation were performed — a direct one, which included manual evaluation done by a human expert, and the indirect one, which included evaluating the performance of CERMINE system trained on GROTOAP2 dataset.
5.1 Manual Evaluation
For the direct manual evaluation we chose a random sample of 50 documents (different than the sample used to construct the rules). We evaluated two document sets: files obtained before applying heuristic-based rules and the same documents from the final dataset. The groups contain 6,228 and 5,813 zones in total, respectively (the difference is related to the zone merging step which reduces the overall number of zones). In both groups the errors were corrected by a human expert, and the original files were compared to the corrected ones, which gave the precision and recall values of the annotation process for each zone label for two stages of the process. The overall accuracy of the annotation process increased from 0.78 to 0.93 after applying heuristic rules. More details about the results of the evaluation can be found in Table 3.
| |
without rules |
with rules |
| precision |
recall |
F1 |
precision |
recall |
F1 |
| abstract |
0.93 |
0.96 |
0.94 |
0.98 |
0.98 |
0.98 |
| acknowledgment |
0.98 |
0.67 |
0.80 |
1.0 |
0.90 |
0.95 |
| affiliation |
0.77 |
0.90 |
0.83 |
0.95 |
0.95 |
0.95 |
| author |
0.85 |
0.95 |
0.90 |
1.0 |
0.98 |
0.99 |
| bib_info |
0.95 |
0.45 |
0.62 |
0.96 |
0.94 |
0.95 |
| body |
0.65 |
0.98 |
0.79 |
0.88 |
0.99 |
0.93 |
| conflict_statement |
0.63 |
0.24 |
0.35 |
0.82 |
0.89 |
0.85 |
| copyright |
0.71 |
0.94 |
0.81 |
0.93 |
0.78 |
0.85 |
| correspondence |
1.0 |
0.72 |
0.84 |
1.0 |
0.97 |
0.99 |
| dates |
0.28 |
1.0 |
0.44 |
0.94 |
1.0 |
0.97 |
| editor |
— |
0 |
— |
1.0 |
1.0 |
1.0 |
| equation |
— |
— |
— |
— |
— |
— |
| figure |
0.99 |
0.36 |
0.53 |
0.99 |
0.46 |
0.63 |
| glossary |
1.0 |
1.0 |
1.0 |
1.0 |
1.0 |
1.0 |
| keywords |
0.94 |
0.94 |
0.94 |
1.0 |
0.94 |
0.97 |
| page_number |
0.99 |
0.53 |
0.69 |
0.98 |
0.97 |
0.98 |
| references |
0.91 |
0.95 |
0.93 |
0.99 |
0.95 |
0.97 |
| table |
0.98 |
0.83 |
0.90 |
0.98 |
0.96 |
0.97 |
| title |
0.51 |
1.0 |
0.67 |
1.0 |
1.0 |
1.0 |
| title_author |
— |
0 |
— |
1.0 |
1.0 |
1.0 |
| type |
0.76 |
0.46 |
0.57 |
0.89 |
0.47 |
0.62 |
| unknown |
0.22 |
0.46 |
0.30 |
0.62 |
0.94 |
0.75 |
| average |
0.79 |
0.68 |
0.73 |
0.95 |
0.91 |
0.92 |
Table 3: The results of manual evaluation of GROTOAP2. The table shows the precision, recall and F1 values for every zone label for the annotation process without and with heuristic-based rules. The correct labels for zones were provided by a human expert. No values appear for labels that were not present in the automatically annotated dataset.
5.2 CERMINE-based Evaluation
For the indirect evaluation we chose randomly a sample of 1000 documents from GROTOAP2 and used them to train CERMINE [8] — our system for extracting metadata and content from scientific publications. CERMINE is able to process documents in PDF format and extracts: document's metadata (including title, authors, emails, affiliations, abstract, keywords, journal name, volume, issue, pages range and year), parsed bibliographic references and the structure of document's sections, section titles and paragraphs. CERMINE is based on a modular workflow composed of a number of steps. Most implementations utilize supervised and unsupervised machine-leaning techniques, which increases the maintainability of the system, as well as its ability to adapt to new document layouts.
The most important parts of CERMINE, that have the strongest impact on the extraction results are: page segmenter and two zone classifiers. Page segmenter recognizes words, lines and zones in the document using Docstrum algorithm [4]. Initial zone classifier labels zones with one of four general categories: metadata, references, body and other. Metadata zone classifier assigns specific metadata classes to metadata zones. Both zone classifiers are based on Support Vector Machines. More details can be found in [8].
We evaluated the overall metadata extraction performance of CERMINE with retrained zone classifiers on 500 PDF documents randomly chosen from PMC. The details of the evaluation methodology can be found in [8]. The results are shown in Table 4. The retrained system obtained a good mean F1 score of 79.34%, which shows the usefulness of GROTOAP2 dataset.
| |
precision |
recall |
F1 |
| title |
93.05% |
88.40% |
90.67% |
| author |
94.38% |
90.01% |
92.14% |
| affiliation |
84.20% |
78.03% |
81.00% |
| abstract |
85.24% |
83.67% |
84.45% |
| keywords |
87.98% |
65.30% |
74.96% |
| journal name |
71.88% |
63.40% |
67.38% |
| volume |
96.28% |
93.20% |
94.72% |
| issue |
49.12% |
55.67% |
52.19% |
| pages |
47.41% |
45.79% |
46.59% |
| year |
99.79% |
97.80% |
98.29% |
| DOI |
96.12% |
85.34% |
90.41% |
| mean |
82.22% |
76.96% |
79.34% |
Table 4: The results of the evaluation of CERMINE system trained on GROTOAP2 dataset. The table shows the precision, recall and F1 values for the extraction of various metadata types, as well as the mean precision, recall and F1 values.
We also compared the overall performance of CERMINE system trained on the entire GROTOAP dataset and randomly chosen 1000 documents from GROTOAP2 dataset in two versions: before applying correction rules and after. The system achieved the average F1 score 62.41% when trained on GROTOAP, 75.38% when trained on GROTOAP2 before applying rules and 79.34% when trained on the final version of GROTOAP2. The results are shown in Table 5.
| |
GROTOAP |
GROTOAP2 |
| without rules |
with rules |
Precision
Recall
F1 |
77.13%
55.99%
62.41% |
81.88%
70.94%
75.38% |
82.22%
76.96%
79.34% |
Table 5: The comparison of the performance of CERMINE trained on GROTOAP, GROTOAP2 before applying improvement rules and final GROTOAP2. The table shows the mean precision, recall and F1 values calculated as an average of the values for individual metadata classes.
6. Conclusions and Future Work
We presented GROTOAP2 — a test set useful for training and evaluation of content analysis-related tasks like zone classification. We described in details the content of the test set and the automatic process of creating it, we also discussed its advantages and drawbacks.
GROTOAP2 contains 13,210 scientific publications in the form of a hierarchical geometric structure. The structure preserves the entire text content of the corresponding PDF documents, geometric features of all the objects, the reading order of the elements, and the labels denoting the role of text fragments in the document.
The method used to create GROTOAP2 is semi-automatic, but does not require manual correction of every document. As a result the method is highly scalable and allows to create large datasets, but the resulting set may contain labelling errors. The manual evaluation of GROTOAP2 showed that the labelling in the dataset is 93% accurate. Despite the errors and thanks to the large volume of GROTOAP2, the dataset is still very useful, which we showed by evaluating the performance of CERMINE system trained on various versions of the dataset.
The main features distinguishing GROTOAP2 from earlier efforts are:
- usefulness in testing algorithms optimized for processing born-digital content,
- reliance on Open Access publications which guarantees easy distribution of both original material and derived ground truth data,
- large volume and the possibility to expand the test set in the future thanks to the semi-automatic scalable creation method.
Our future plans include:
- generating a dataset of parsed bibliographic references by extracting reference strings from PDF files and labelling their fragments based on NLM data,
- assigning more specific body labels, especially for section headers of different levels,
- enriching the ground truth files with the names of the fonts extracted from PDF files.
Acknowledgements
This work has been partially supported by the European Commission as part of the FP7 project OpenAIREplus (grant no. 283595).
References
[1] A. Antonacopoulos, D. Bridson, C. Papadopoulos, and S. Pletschacher. A Realistic Dataset for Performance Evaluation of Document Layout Analysis. 2009 10th International Conference on Document Analysis and Recognition, pages 296—300, 2009. http://doi.org/10.1109/ICDAR.2009.271
[2] C. H. Lee and T. Kanungo. The architecture of TrueViz: a groundTRUth/metadata editing and VIsualiZing ToolKit. Pattern Recognition, 15, 2002. http://doi.org/10.1016/S0031-3203(02)00101-2
[3] D. Lewis, G. Agam, S. Argamon, O. Frieder, D. Grossman, and J. Heard. Building a Test Collection for Complex Document Information Processing. In Proc. 29th Annual Int. ACM SIGIR Conference, pages 665-666, 2006. http://doi.org/10.1145/1148170.1148307
[4] L. O'Gorman. The document spectrum for page layout analysis. IEEE Transactions on Pattern Analysis and Machine Intelligence, 15(11):1162—1173, 1993. http://doi.org/10.1109/34.244677
[5] J. Sauvola and H. Kauniskangas. MediaTeam Document Database II, a CD-ROM collection of document images, University of Oulu, Finland, 1999.
[6] T. Smith and M. Waterman. Identification of common molecular subsequences. Journal of Molecular Biology, 147(1):195—197, 1981. http://doi.org/10.1016/0022-2836(81)90087-5
[7] D. Tkaczyk, A. Czeczko, K. Rusek, L. Bolikowski, and R. Bogacewicz. Grotoap: ground truth for open access publications. In 12th ACM/IEEE-CS Joint Conference on Digital Libraries, pages 381—382, 2012. http://doi.org/10.1145/2232817.2232901
[8] D. Tkaczyk, P. Szostek, P. J. Dendek, M. Fedoryszak, and L. Bolikowski. CERMINE - automatic extraction of metadata and references from scientific literature. In Proceedings of the 11th IAPR International Workshop on Document Analysis Systems, pages 217—221, 2014. http://doi.org/10.1109/DAS.2014.63
About the Authors
 |
Dominika Tkaczyk is a researcher at the Interdisciplinary Centre for Mathematical and Computational Modelling at University of Warsaw (ICM UW). She received her MSc in Computer Science from University of Warsaw. Her current research interests focus on extensive analysis of scientific literature. She is the lead developer of CERMINE - a machine learning-based Java library for extracting metadata and content from scholarly publications. She has also contributed to the design and development of a big data knowledge discovery service for OpenAIREplus project. Previously she was involved in developing YADDA2 system — a scalable open software platform for digital library applications.
|
 |
Pawel Szostek is a computer scientist working as a Fellow in CERN openlab, Switzerland. He is focused on profiling High Energy Physics applications, benchmarking, efficient computing and performance monitoring. His duties encompass advanced computing studies using various machines and compilers, as well as managing a cluster of servers used for software development for experiments at CERN. He graduated in Computer Science from Warsaw University of Technology. Previously he was employed at the University of Warsaw where he focused on machine learning techniques applied to immense sets of scholarly articles gathered in a digital library.
|
 |
Łukasz Bolikowski is an Assistant Professor at the Interdisciplinary Centre for Mathematical and Computational Modelling at University of Warsaw (ICM UW). He defended his PhD thesis on semantic network analysis at Systems Research Institute of Polish Academy of Sciences. He is a leader of a research group focusing on scalable knowledge discovery in scholarly publications. He has contributed to a number of European projects, including DRIVER II, EuDML, OpenAIREplus. Earlier at ICM UW he specialized in construction of mathematical models and their optimization on High Performance Computing architectures.
|
|